The proliferative history shapes the DNA methylome of B-cell tumors and predicts clinical outcome
Author/s: Martí Duran-Ferrer, Guillem Clot, Ferran Nadeu, Renée Beekman, Tycho Baumann, Jessica Nordlund, Yanara Marincevic-Zuniga, Gudmar Lönnerholm, Alfredo Rivas-Delgado, Silvia Martin, Raquel Ordoñez, Giancarlo Castellano, Marta Kulis, Ana Queirós, Lee Seung-Tae, Joseph Wiemels, Romina Royo, Montserrat Puiggrós, Junyan Lu, Eva Gine, Sílvia Beà, Pedro Jares, Xabier Agirre, Felipe Prosper, Carlos López-Otín, Xosé S.Puente, Christopher C. Oakes, Thorsten Zenz, Julio Delgado, Armando López-Guillermo, Elías Campo and José Ignacio Martin-Subero
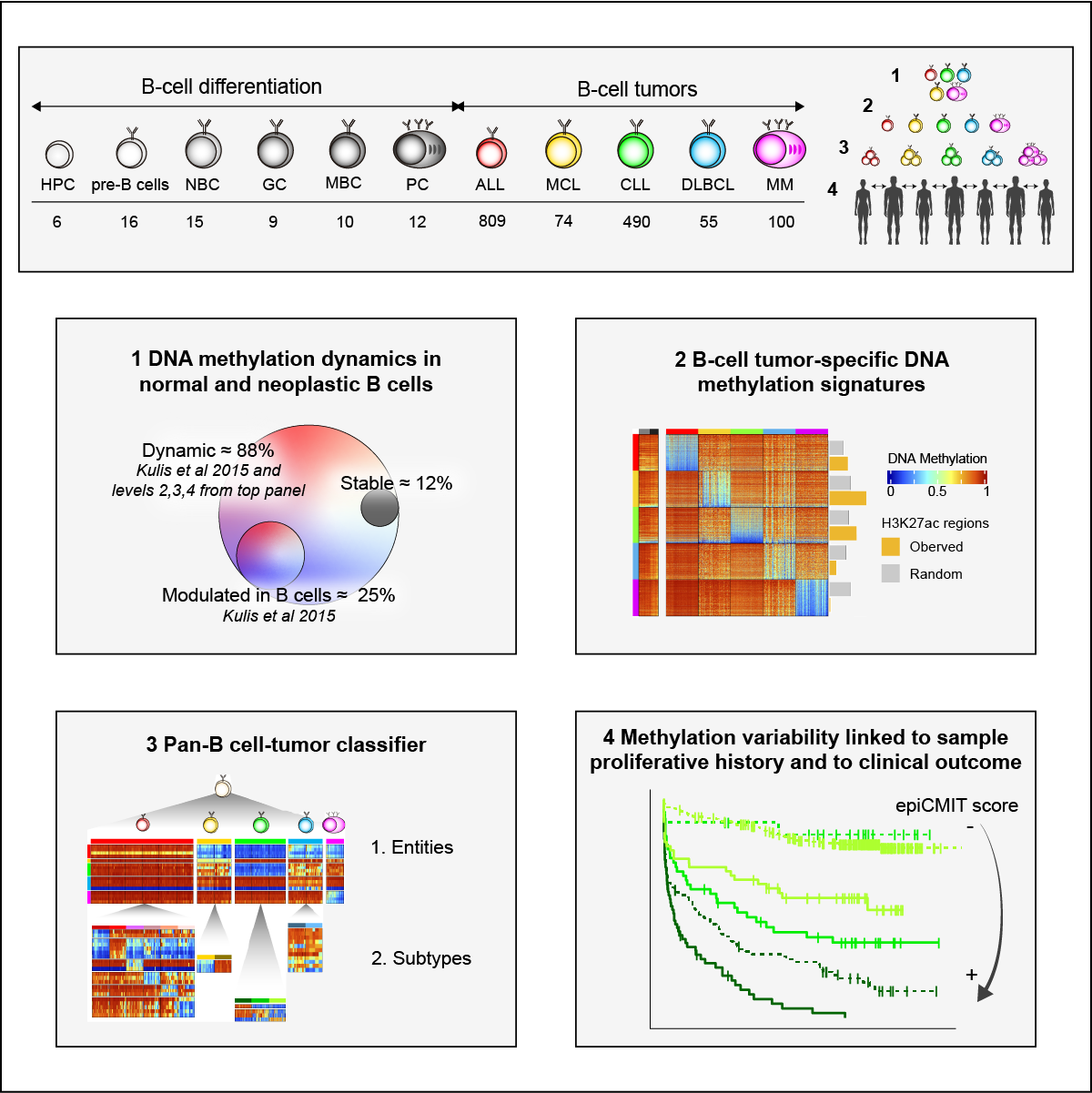
Abstract
We report a systematic analysis of the DNA methylation variability in 1,595 samples of normal cell subpopulations and 14 tumor subtypes spanning the entire human B-cell lineage. Differential methylation among tumor entities relates to differences in cellular origin and to de novo epigenetic alterations, which allowed us to build an accurate machine learning-based diagnostic algorithm. We identify extensive patient-specific methylation variability in silenced chromatin associated with the proliferative history of normal and neoplastic B cells. Mitotic activity generally leaves both hyper- and hypomethylation imprints, but some B-cell neoplasms preferentially gain or lose DNA methylation. Subsequently, we construct a DNA methylation-based mitotic clock called epiCMIT, whose lapse magnitude represents a strong independent prognostic variable in B-cell tumors and is associated with particular driver genetic alterations. Our findings reveal DNA methylation as a holistic tracer of B-cell tumor developmental history, with implications in the differential diagnosis and prediction of clinical outcome.Graphical summary
Data availablility
| Description | File size | Download |
| DNA methylation matrix for 1799 samples and related metadata | 6.1G | Link |
| DNA methylation of curated 1595 samples and related metadata | 4.9G | Link |
| DNA methylation for CLL validation series and related metadata | 1.8G | Link |
| DNA methlation for MCL validation series and related metadata | 465M | Link |
| DNA methylation for DLBCL validation series and related metadata | 137M | Link |
Files are compressed and password protected. Please, contact either Martí Duran-Ferrer (maduran@clinic.cat) or Iñaki Martín-Subero (imartins@clinic.cat) to obtain the password.
Code availability
The source code is available at https://github.com/Duran-FerrerM/Pan-B-cell-methylome