The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia
Author/s: Renée Beekman, Vicente Chapaprieta, Núria Russiñol, Roser Vilarrasa-Blasi, Núria Verdaguer-Dot, Joost H.A. Martens, Martí Duran-Ferrer, Marta Kulis, François Serra, Biola M. Javierre, Steven W. Wingett, Guillem Clot, Ana C. Queirós, Giancarlo Castellano, Julie Blanc, Marta Gut, Angelika Merkel, Simon Heath, Anna Vlasova, Sebastian Ullrich, Emilio Palumbo, Anna Enjuanes, David Martín-García, Sílvia Beà, Magda Pinyol, Marta Aymerich, Romina Royo, Montserrat Puiggros, David Torrents, Avik Datta, Ernesto Lowy, Myrto Kostadima, Maša Roller, Laura Clarke, Paul Flicek, Xabier Agirre, Felipe Prosper, Tycho Baumann, Julio Delgado, Armando López-Guillermo, Peter Fraser, Marie-Laure Yaspo, Roderic Guigó, Reiner Siebert, Marc A. Martí-Renom, Xose S. Puente, Carlos López-Otín, Ivo Gut, Hendrik G. Stunnenberg, Elias Campo & Jose I. Martin-Subero
Nature Medicine, volume 24, pages 868–880 (2018)
doi: 10.1038/s41591-018-0028-4
Abstract
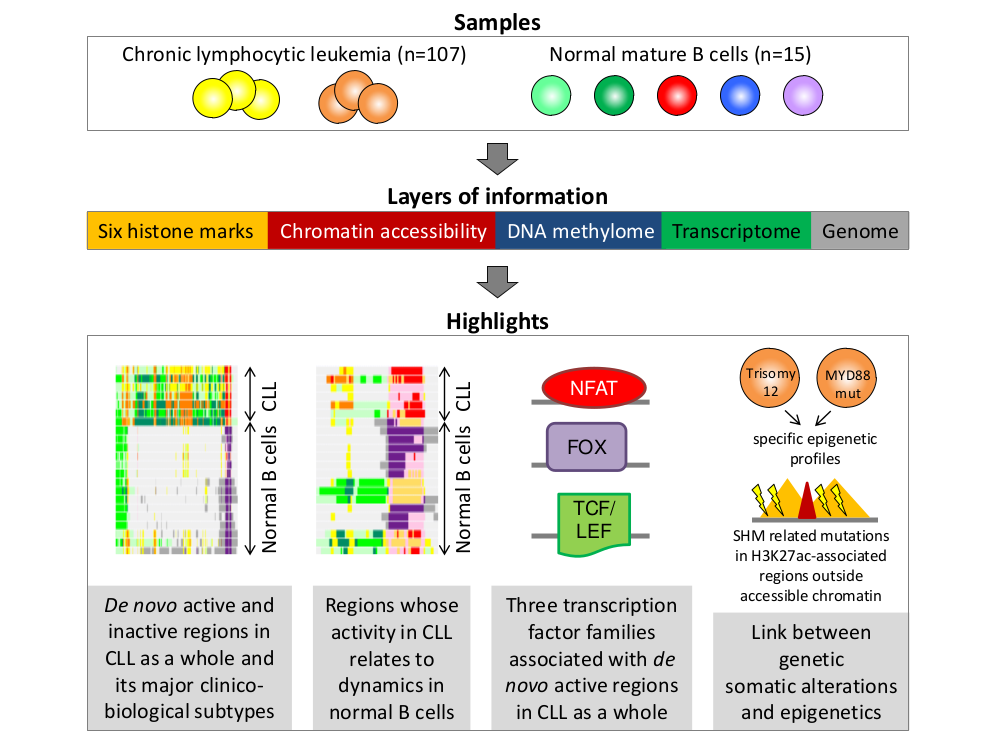
Chronic lymphocytic leukemia (CLL) is a frequent hematological neoplasm in which underlying epigenetic alterations are only partially understood. Here, we analyze the reference epigenome of seven primary CLLs and the regulatory chromatin landscape of 107 primary cases in the context of normal B cell differentiation. We identify that the CLL chromatin landscape is largely influenced by distinct dynamics during normal B cell maturation. Beyond this, we define extensive catalogues of regulatory elements de novo reprogrammed in CLL as a whole and in its major clinico-biological subtypes classified by IGHV somatic hypermutation levels. We uncover that IGHV-unmutated CLLs harbor more active and open chromatin than IGHV-mutated cases. Furthermore, we show that de novo active regions in CLL are enriched for NFAT, FOX and TCF/LEF transcription factor family binding sites. Although most genetic alterations are not associated with consistent epigenetic profiles, CLLs with MYD88 mutations and trisomy 12 show distinct chromatin configurations. Furthermore, we observe that non-coding mutations in IGHV-mutated CLLs are enriched in H3K27ac-associated regulatory elements outside accessible chromatin. Overall, this study provides an integrative portrait of the CLL epigenome, identifies extensive networks of altered regulatory elements and sheds light on the relationship between the genetic and epigenetic architecture of the disease.Graphical summary
Additional information
UCSC tracks
Please, use this link to access the CLL Referece Epigenome tracks in the UCSC genome browser. The presented tracks are briefly described in this document.
Data
All the raw data (FASTQ and BAM files) included in this study have been deposited and released, as part of the BLUEPRINT epigenome project, at the European Genome-Phenome Archive (EGA, https://ega-archive.org). They can be found under the unifying EGA accession number EGAD00001004046.
Additional results can be found in the next table.
| File | Details | File size | Download |
|
CLL & B cells chromatin states |
Assignment of 12 different chromatin states in 200 base pair bins (GRCh38) using the chromHMM software |
52M | Link |
|
Normalized signal intensities of CLL reference epigenome & B cells for RNA-seq, H3K27ac, H3K4me1, H3K4me3, H3K9me3, H3K27me3, H3K36me3 & ATAC-seq |
Variance stabilizing transformed data signals as determined using DEseq2 in consensus peaks (GRCh38) for the different histone marks and ATAC-seq.
FPKM gene expression values for RNA-seq (gencode22). |
43M | Link |
|
CLL reference epigenome & B cells methylation |
DNA methylation estimates (GRCh38) determined by whole genome bisulfite sequencing. |
431M | Link |
|
Normalized signal intensities of CLL extended dataset & B cells for RNA-seq, H3K27ac& ATAC-seq |
Variance stabilizing transformed data signals as determined using DEseq2 in consensus peaks (GRCh38) for H3K27ac and ATAC-seq, corrected for the SPOT score.
FPKM gene expression values for RNA-seq (gencode22). |
227M | Link |
| TADs GM12878 |
Topologically associating domains (GRCh38) in the lymphoblastoid cell line GM12878 as determined by TADbit using Hi-C data generated by Rao et al. (PMID 25497547) |
25K | Link |
|
Interactions Promoter capture Hi-C |
Three dimensional chromatin interactions in CLL, naive B cells (NB) and total B cells from peripheral blood (TB_merged). For the latter 2, the data generated by Javierre et al. was used (PMID 27863249). For each interaction the bait (first 3 columns) and the interacting fragment (column 4-6) are indicated (GRCh38) as well as the interaction score determined by the CHICAGO software (column 7). |
6M | Link |